Jakarta Apartment Rental Estimates
During the holiday I decided to make this regression project which try to estimate the prices of apartment rental in Jakarta. I know that there’s a lot of these type of property prediction project out there, especially on Kaggle. So I try to make it different by scraping my own data from the Travelio website. Overall, this is a fun experience for me I got to learn a lot of new things like linear regression assumption, web scraping with Selenium and Beautiful Soup, geocoding with google Map API, and trying out various ensemble learning methods.
Overview
- A tool that estimate monthly apartment rental prices in Jakarta. This could tool be use to help negotiate rental prices when finding an apartment to stay in Jakarta.
- Scraped over 800 data of apartment in Jakarta using selenium on travelio.com website.
- Perform cleaning on some columns: Integer extraction, remove row with missing value, iterative imputing with Bayesian Ridge, and removing outlier.
- EDA using various plot like distribution plot, histogram, scatter plot, heatmap, and prob plot.
- Perform geocoding using Google Maps API; extract coordinates from addresses and plot their price heatmap distribution.
- Various feature engineering like removing multicollinear features, encoding categorical features, log transform skewed features, and parsing out address to create new sub-district/kecamatan feature.
- Optimized Linear, Ridge, Random Forest, Gradient Boosing, and XGBoost model using methods like Random Search CV and manually trying different parameters.
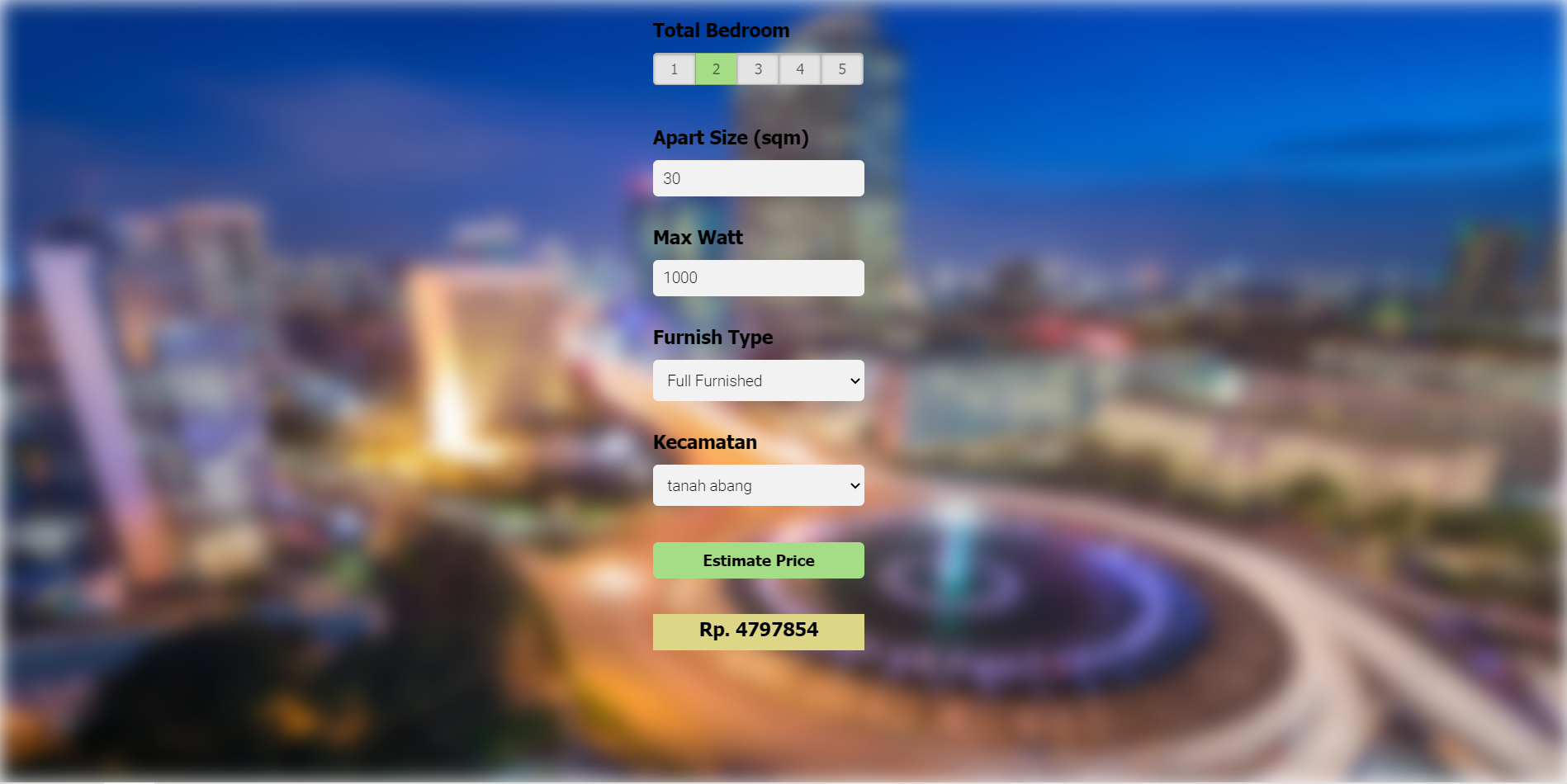
- Built a client facing API using flask and a simple website interface with html,css, js to simulate how users can input apartment data and get their rental price estimate.
Web Scraping
Since the code for the web scrapping is quite long I’ll just attach a link that leads to it. The scraping result is stored in the travelio.csv file.
df = pd.read_csv('travelio.csv', index_col = 0)
Data Cleaning
print(df.isna().sum())
print(f"Number of column before cleaning: {df.shape[0]}")
Name 0
Total Bedroom 0
Total Bathroom 0
Apart Size 0
Max Capacity 33
Max Watt 28
Address 33
Swim Pool 0
Rating 0
Total Review 23
Furnish Type 0
Price 0
dtype: int64
Number of column before cleaning: 806
It seems that there are some rows that are missing some important informations. Since it will be really hard to fill in those data with appropriate value we will just remove them instead.
# Remove rows that have no address
df2 = df.dropna(subset = ['Address'])
# Because our job is to predict apartment prices from the given specifications it wouldn't make sense to actually include 'Rating' and
# 'Total Review' since those variables can only be obtained when the apartment is already listed along with its price.
df2.drop(columns = ['Rating', 'Total Review'], inplace = True)
print(df2.isna().sum())
print(f"Number of column after cleaning: {df2.shape[0]}")
Name 0
Total Bedroom 0
Total Bathroom 0
Apart Size 0
Max Capacity 0
Max Watt 0
Address 0
Swim Pool 0
Furnish Type 0
Price 0
dtype: int64
Number of column after cleaning: 773
df2.reset_index(drop=True, inplace=True)
# Clean the price column from the dataset and extract only the integer
df2['Price'] = df2['Price'].str.replace(r'\D+', '')
df2['Price'] = df2['Price'].astype(int)
df2['Apart Size'] = df2['Apart Size'].str.replace(r'\D+', '')
df2['Apart Size'] = df2['Apart Size'].astype(int)
df2['Max Capacity'] = df2['Max Capacity'].str.replace(r'\D+', '')
df2['Max Capacity'] = df2['Max Capacity'].astype(int)
df2['Max Watt'] = df2['Max Watt'].str.replace(r'\D+', '')
df2['Max Watt'] = df2['Max Watt'].astype(int)
df2['Total Bedroom'].replace({"Studio": 0}, inplace = True)
df2['Total Bedroom'] = df2['Total Bedroom'].astype(int)
df2.info()
df2.describe()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 773 entries, 0 to 772
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Name 773 non-null object
1 Total Bedroom 773 non-null int32
2 Total Bathroom 773 non-null int64
3 Apart Size 773 non-null int32
4 Max Capacity 773 non-null int32
5 Max Watt 773 non-null int32
6 Address 773 non-null object
7 Swim Pool 773 non-null int64
8 Furnish Type 773 non-null object
9 Price 773 non-null int32
dtypes: int32(5), int64(2), object(3)
memory usage: 45.4+ KB
| Total Bedroom | Total Bathroom | Apart Size | Max Capacity | Max Watt | Swim Pool | Price | |
|---|---|---|---|---|---|---|---|
| count | 773.000000 | 773.000000 | 773.000000 | 773.000000 | 773.000000 | 773.000000 | 7.730000e+02 |
| mean | 1.349288 | 1.179819 | 51.058215 | 2.915912 | 2545.725744 | 0.878396 | 6.816667e+06 |
| std | 0.989382 | 0.413511 | 32.613103 | 1.064947 | 2016.470858 | 0.478243 | 4.224733e+06 |
| min | 0.000000 | 1.000000 | 14.000000 | 1.000000 | 1.000000 | -1.000000 | 1.799999e+06 |
| 25% | 0.000000 | 1.000000 | 31.000000 | 2.000000 | 1300.000000 | 1.000000 | 4.199391e+06 |
| 50% | 2.000000 | 1.000000 | 38.000000 | 3.000000 | 2200.000000 | 1.000000 | 5.281957e+06 |
| 75% | 2.000000 | 1.000000 | 61.000000 | 4.000000 | 3500.000000 | 1.000000 | 8.206249e+06 |
| max | 3.000000 | 4.000000 | 297.000000 | 8.000000 | 21000.000000 | 1.000000 | 3.777713e+07 |
EDA
sb.distplot(df2['Price'])
<AxesSubplot:xlabel='Price', ylabel='Density'>

print("Skewness: %f" % df2['Price'].skew())
print("Kurtosis: %f" % df2['Price'].kurt())
Skewness: 1.993154
Kurtosis: 5.708796
Skewness can indicate wether the data follow the normal distribution, in most cases a skewed data can actually lead to a worst performance of the model. Most model like linear regression works better with a data that follows the normal distribution.
On the other hand, kurtosis indicate how heavy or light tailed the data is which is correlated to the presence of outliers. A kurtosis higher than 3 (normal distribution) normally indicate the presence of outliers and have a very peaked shape.
sb.histplot(data = df2['Apart Size'] )
<AxesSubplot:xlabel='Apart Size', ylabel='Count'>

print("Skewness: %f" % df2['Apart Size'].skew())
print("Kurtosis: %f" % df2['Apart Size'].kurt())
Skewness: 2.195653
Kurtosis: 7.163900
sb.pairplot(df2)
plt.rcParams["figure.figsize"] = [16,9]
plt.show()

Based on our scatter plot there is sign of outliers in Apart Size, we will investigate it further along the line.
fig, ax = plt.subplots(figsize=(16, 9))
corr_matrix = df2.corr()
sb.heatmap(data = corr_matrix, annot = True)
plt.show()

The heatmap shows sign of multicollinearity between ‘Total Bedroom’, ‘Total Bathroom’, ‘Apart Size’, and ‘Max Capacity’. We many need to remove some of these variables. The heatmap also shows a strong linear correlation between our target variable ‘Price’ and ‘Apart Size’ so its definitely a feature we want to keep.
# Checkpoint
# df2.to_csv('travelio2.csv', index=True)
df2 = pd.read_csv('travelio2.csv', index_col = 0)
Geocoding
import geopy
from geopy.extra.rate_limiter import RateLimiter
from geopy.exc import GeocoderTimedOut
from geopy.geocoders import GoogleV3
from geopy.extra.rate_limiter import RateLimiter
geolocator = GoogleV3(api_key = '***************')
# Delay 1 second between each call to reduce the probability of a time out
geocode = RateLimiter(geolocator.geocode, min_delay_seconds=1)
df2['location'] = df2['Address'].apply(geocode)
df2['point'] = df2["location"].apply(lambda loc: tuple(loc.point) if loc else None)
# Unload the points to latitude and longtitude
df2[['lat', 'lon', 'altitude']] = pd.DataFrame(df2['point'].to_list(), index=df2.index)
Now that we have longitude and altitude of each address we don’t need location and point features anymore.
We should also remove altitude because Google API require us to use the latitude api if we want to get latitude for a specific location. We won’t go that far since its unlikely that altitude will play any significant roles on Jakarta apartments rental prices.
df2.drop(columns = ['point', 'altitude'], inplace = True)
# Checkpoint
# df2.to_csv('travelio3.csv', index=True)
df2 = pd.read_csv('travelio3.csv', index_col = 0)
gmaps.configure(api_key='*********')
fig = gmaps.figure()
heatmap_layer = gmaps.heatmap_layer(
df2[['lat','lon']],
weights= df2['Price'],
max_intensity = 100000000,
point_radius= 10.0
)
fig.add_layer(heatmap_layer)
fig

Based on our geographical analysis we can see that apartment prices tend to be higher when located in Central and South Jakarta region. This is as expected as those 2 region are known as elite places in Jakarta with lots of facilities and well-developed infrastructures.
Feature Engineering
Multicollinearity
Since we are going to run a multiple linear regression model on this dataset it is important to avoid multicollinearity. When there is a correlation between the independent variables it can becomes difficult for the model to estimate the relationship between each independent variable and the dependent variable independently because the independent variables tend to change in unison.
Here are some algorithms that are effected by multicollinearity Linear Regression, Logistic Regression, KNN, and Naive Bayes.
It seems that ‘Max Capacity’ has a strong correlation with total Bedroom which indicate some multicollinearity. If we think about it, it makses sense because max capacity of an apartment is probably based on the number of bedroom and size of apartment.
df2.drop(columns = "Max Capacity", inplace = True)
X_variables = df2[['Total Bedroom', 'Total Bathroom', 'Apart Size', 'Max Watt']]
vif_data = pd.DataFrame()
vif_data["feature"] = X_variables.columns
vif_data["VIF"] = [variance_inflation_factor(X_variables.values, i) for i in range(len(X_variables.columns))]
vif_data
| feature | VIF | |
|---|---|---|
| 0 | Total Bedroom | 4.468788 |
| 1 | Total Bathroom | 9.512638 |
| 2 | Apart Size | 10.778528 |
| 3 | Max Watt | 3.888674 |
As seen on the heatmap and vif Total Bathroom and Apart Size seems highly correlated so we can remove one of them. I decided to remove Total Bathroom as it has a lower correlation with the target variable ‘Price’.
df2.drop(columns = "Total Bathroom", inplace = True)
X_variables = df2[['Total Bedroom','Apart Size', 'Max Watt']]
vif_data = pd.DataFrame()
vif_data["feature"] = X_variables.columns
vif_data["VIF"] = [variance_inflation_factor(X_variables.values, i) for i in range(len(X_variables.columns))]
vif_data
| feature | VIF | |
|---|---|---|
| 0 | Total Bedroom | 4.280758 |
| 1 | Apart Size | 6.205461 |
| 2 | Max Watt | 3.688159 |
# Checkpoint
# df2.to_csv('travelio4.csv', index = True)
df2 = pd.read_csv('travelio4.csv', index_col = 0)
Get Sub-Districts
I will try to obtain the list of districts and sub-districts each apartment is located in. This might help with our prediction since we now that some districts are more elite than others, these elite districts tend to have higher property prices.
import requests
from bs4 import BeautifulSoup
URL = "https://id.wikipedia.org/wiki/Daftar_kecamatan_dan_kelurahan_di_Daerah_Khusus_Ibukota_Jakarta"
page = requests.get(URL)
soup = BeautifulSoup(page.content, "html.parser")
table = soup.find_all("tbody")
kecamatan = []
element = soup.select("td:nth-of-type(2)")
for idx, item in enumerate(element):
if idx >5 and idx <48:
kecamatan.append(item.text)
old = ['Kelapa Gading', 'Pasar Minggu', 'Pasar Rebo', 'Tanjung Priok', 'Kebayoran Lama', 'Kebayoran Baru', 'Mampang Prapatan', 'Kebon Jeruk']
new = ['Klp. Gading', 'Ps. Minggu', 'Ps. Rebo', 'Tj. Priok', 'Kby. Lama', 'Kby. Baru','Mampang Prpt', 'Kb. Jeruk']
for i, val in enumerate(old):
idx = kecamatan.index(val)
kecamatan[idx] = new[i]
kecamatan.append('Setia Budi')
kecamatan.append('Pd. Aren')
kecamatan.append('Kebon Jeruk')
# Create a function that will return the district an address belongs in, and apply that function to every row in the data
def kec(val):
return next((x for x in kecamatan if x.lower() in val.lower()), None)
df2['kecamatan'] = df2['location'].apply(kec)
a = df2[df2['kecamatan'].isnull()].index.tolist()
empty_rows = df2.iloc[a].copy()
print(a)
[96, 97, 110, 164, 172, 190, 233, 235, 240, 243, 249, 573, 627, 641, 646, 668, 682, 688, 689, 743]
It seems that some locations are incomplete (they don’t have district in them) due to the geocoding by Google API, so we will just use our original Address that we obtain from the travelio website instead.
rev = testdf.index.to_list()
def kec(val):
return next((x for x in kecamatan if x.lower() in val.lower()), None)
df2['kecamatan'].iloc[rev] = df2['Address'].iloc[rev].apply(kec)
df2.replace({'Setia Budi' : 'Setiabudi'}, inplace=True)
df2.replace({'Kebon Jeruk' : 'Kb. Jeruk'}, inplace=True)
a= df2[df2['kecamatan'].isnull()].index.tolist()
len(df2.iloc[a])
0
Great! We’ve filled in all the districts for each corresponding apartment. Now lets try to see the average price of housing for each district.
df2.groupby(['kecamatan']).Price.agg('mean').sort_values(ascending = False)
# res = df_agg.apply(lambda x: x.sort_values(ascending=True))
kecamatan
Kby. Baru 1.806751e+07
Mampang Prpt 1.523673e+07
Setiabudi 1.106564e+07
Kby. Lama 1.089827e+07
Menteng 9.563951e+06
Cilandak 8.927184e+06
Pancoran 8.576855e+06
Tanah Abang 7.904778e+06
Pd. Aren 7.735000e+06
Gambir 6.446666e+06
Kb. Jeruk 6.430905e+06
Pesanggrahan 6.270067e+06
Tebet 5.843505e+06
Grogol Petamburan 5.749463e+06
Senen 5.660513e+06
Kembangan 5.596787e+06
Kemayoran 5.463723e+06
Tambora 5.285560e+06
Kalideres 5.280624e+06
Ps. Minggu 5.273301e+06
Pademangan 5.096884e+06
Kramat Jati 4.981074e+06
Penjaringan 4.865202e+06
Jatinegara 4.780318e+06
Klp. Gading 4.778076e+06
Sawah Besar 4.761854e+06
Ps. Rebo 4.500187e+06
Taman Sari 4.375700e+06
Cengkareng 4.273109e+06
Cempaka Putih 4.173814e+06
Pulo Gadung 3.681220e+06
Tj. Priok 3.512187e+06
Duren Sawit 2.959583e+06
Cakung 2.623194e+06
Name: Price, dtype: float64
As we can see, the top 5 districts with the highest apartment price in average comes from district located in South and Central Jakarta. This make sense since we know that places in South and Central Jakarta have a higher living cost and are more elite compare to other region.
A bit more cleaning
While I was extracting district for each row I’ve also discovered that some of the Max Watt have only a value of 1, it seems that I forgot to clean these rows. These 1’s indicate that the scraper are unable to extract the max watt information during the web scraping process.
So, I’ll have to perform some imputing. To impute this data I’ve decided to use multivariate imputing from Sk-Learn which uses a Bayesian Ridge Regression to predict the most likely outcome of the missing data using other features in the data. We basically treat Max Watt as the dependent variables.
This should result to a more accurate imputation compared to the univariate imputation in which only a single value obtain by calculating the mean or median is use to fill in all the missing data.
# Checkpoints
# df2.to_csv('travelio5.csv', index = True)
df2 = pd.read_csv('travelio5.csv', index_col=0)
df2['Max Watt'] = df2['Max Watt'].replace(1, np.NaN)
df2['Max Watt'].isna().sum()
66
# Here I created a temporary dataframe and fill it with features that have highest correlations with Max Watt.
# These features will act as the independent/predictors variable to determine the most likely value for Max Watt.
# I pick these features as predictor because they have the highest correlation with Max Watt
dftemp = df2[['Total Bedroom', 'Apart Size', 'Max Watt', 'Price']].copy()
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
from sklearn.linear_model import BayesianRidge
imputer = IterativeImputer()
imputer.fit(dftemp)
df_trans = imputer.transform(dftemp)
df_trans = pd.DataFrame(df_trans, columns=dftemp.columns)
df2['Max Watt'] = df_trans['Max Watt']
df2['Max Watt'] = df2['Max Watt'].astype(int)
# Checkpoints
# df2.to_csv('travelio6.csv', index = True)
df2 = pd.read_csv('travelio6.csv', index_col = 0)
Outliers
fig, ax = plt.subplots(figsize=(8, 6))
ax.scatter(x = df2['Apart Size'], y = df2['Price'])
plt.ylabel('Price', fontsize=13)
plt.xlabel('Apart Size', fontsize=13)
plt.show()

We can see that there is one apartment with a very high price despite its fairly average size, we can assume that it is located in a very elite location but unfortunately is simply way to high and unlikely in a real life scenario so we can simply treat it as an outlier and remove that data point.
# Delete Outliers
df2 = df2.drop(df2[df2['Price'] > 35000000].index)
fig, ax = plt.subplots(figsize=(8, 6))
ax.scatter(x = df2['Apart Size'], y = df2['Price'])
plt.ylabel('Price', fontsize=13)
plt.xlabel('Apart Size', fontsize=13)
plt.show()

Handling Categorical Data
df2.reset_index(drop=True, inplace=True)
# Clean some row
df2['Furnish Type'] = df2['Furnish Type'].replace('5', "Unfurnished")
# Dummy Encoding Furnish Type because it is a nominal variable
dummy_furnished = pd.get_dummies(df2[['Furnish Type', 'kecamatan']], prefix='', prefix_sep='')
df2 = pd.merge(
left=df2,
right=dummy_furnished,
left_index=True,
right_index=True,
)
df2.drop(columns = ["Furnish Type", 'kecamatan'], inplace = True)
Normality
To check for the distribution and normality of our features I’ll be plotting a distribution plot and a normal probability plot / Q-Q plot.
Features with normal distribution should have a symemtrical bell shape curve in the distribution plot and data distribution should closely follow the diagonal in the Q-Q plot.
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20,6))
sb.distplot(df2['Price'], fit=norm, ax=ax1)
res = stats.probplot(df2['Price'], plot=ax2)

After removing the outliers in ‘Sales Price’ we can see that the kurtosis becomes closer to that of a normal distribution. However we can still see some skewness in our data so we can fix it using box cox transformation. We will also apply it to “apart size”.
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20,6))
sb.distplot(df2['Apart Size'], fit=norm, ax=ax1)
res = stats.probplot(df2['Apart Size'], plot=ax2)

We can analyze that the Apartment rental pricing are skewed to the left and has a heavy tail distribution.
Log Transform
To fix the skewness and transform our data into a more normal distribution I’m going to perform a log transformation. The reason I did not go with box cox transformation is because it will be difficult to reverse transform our price estimate during the final production.
df2['Apart Size'] = np.log(df2['Apart Size'])
df2['Price'] = np.log(df2['Price'])
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20,6))
sb.distplot(df2['Price'], fit=norm, ax=ax1)
res = stats.probplot(df2['Price'], plot=ax2)

fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20,6))
sb.distplot(df2['Apart Size'], fit=norm, ax=ax1)
res = stats.probplot(df2['Apart Size'], plot=ax2)

Testing Homoscedasticity
Homoscedasticity refers to the ‘assumption that dependent variable(s) exhibit equal levels of variance across the range of predictor variable(s)’. Homoscedasticity is desirable because we want the error term to be the same across all values of the independent variables.
The best approach to test homoscedasticity for two metric variables is graphically. Departures from an equal dispersion are shown by such shapes as cones (small dispersion at one side of the graph, large dispersion at the opposite side) or diamonds (a large number of points at the center of the distribution).
plt.subplots(figsize=(8, 6))
plt.scatter(df2['Price'], df2['Apart Size'])
<matplotlib.collections.PathCollection at 0x27361563e80>

As you can see the scatter plot doesn’t have a conic shape anymore. Thats the power of normality. Just by ensuring normality in some variables, we can ensure homoscedasticity.
fig, ax = plt.subplots(figsize=(16, 9))
corr_matrix = df2.iloc[:, :12].corr()
sb.heatmap(data = corr_matrix, annot = True)
plt.show()

# Checkpoint
# df2.to_csv('travelio7.csv', index=True)
df2 = pd.read_csv('travelio7.csv', index_col=0)
Feature Selection
df2.head()
X_var = df2.drop(columns= ['Name', 'lat', 'lon', 'Swim Pool', 'location', 'Address', 'Price'])
y = df2['Price']
X_var.describe()
| Total Bedroom | Apart Size | Max Watt | Full Furnished | Unfurnished | Cakung | Cempaka Putih | Cengkareng | Cilandak | Duren Sawit | ... | Ps. Rebo | Pulo Gadung | Sawah Besar | Senen | Setiabudi | Taman Sari | Tambora | Tanah Abang | Tebet | Tj. Priok | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | ... | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 | 772.000000 |
| mean | 1.347150 | 3.780989 | 2796.141192 | 0.922280 | 0.077720 | 0.007772 | 0.007772 | 0.025907 | 0.009067 | 0.003886 | ... | 0.005181 | 0.069948 | 0.023316 | 0.036269 | 0.081606 | 0.006477 | 0.003886 | 0.068653 | 0.012953 | 0.012953 |
| std | 0.988234 | 0.519492 | 1881.756917 | 0.267904 | 0.267904 | 0.087873 | 0.087873 | 0.158960 | 0.094851 | 0.062257 | ... | 0.071841 | 0.255225 | 0.151003 | 0.187081 | 0.273941 | 0.080269 | 0.062257 | 0.253027 | 0.113147 | 0.113147 |
| min | 0.000000 | 2.639057 | 900.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 0.000000 | 3.433987 | 1917.250000 | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 50% | 2.000000 | 3.637586 | 2200.000000 | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 75% | 2.000000 | 4.110874 | 3500.000000 | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| max | 3.000000 | 5.693732 | 21000.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | ... | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
8 rows × 39 columns
Train Test Split
I’m going to split the data into a test set and a training set. I will hold out the test set until the very end and use the error on those data as an unbiased estimate of how my models did.
I might perform a further split later on the training set into training set proper and a validation set or I might cross-validate.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_var, y, test_size=0.2, random_state=27)
2. Model
2.0 Cross-Validation Routine
kf = KFold(n_splits=5, shuffle=True, random_state=27)
from sklearn.model_selection import KFold
# squared_loss
def rmse_cv(model):
rmse = -cross_val_score(model, X_train, y_train, scoring="neg_root_mean_squared_error", cv = kf)
return(rmse)
def plot_learning_curve(estimator, title, X, y, axes=None, ylim=None, cv=ShuffleSplit(n_splits=5, test_size=0.2, random_state=0), n_jobs=None, train_sizes=np.linspace(.1, 1.0, 5)):
cv = ShuffleSplit(n_splits=5, test_size=0.2, random_state=27)
if axes is None:
_, axes = plt.subplots(figsize=(10, 8))
axes.set_title(title)
if ylim is not None:
axes.set_ylim(*ylim)
axes.set_xlabel("Training examples")
axes.set_ylabel("Score")
train_sizes, train_scores, test_scores, fit_times, _ = \
learning_curve(estimator, X, y, cv=cv, n_jobs=n_jobs,
train_sizes=train_sizes,
return_times=True)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
fit_times_mean = np.mean(fit_times, axis=1)
fit_times_std = np.std(fit_times, axis=1)
# Plot learning curve
axes.grid()
axes.fill_between(train_sizes, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.1,
color="r")
axes.fill_between(train_sizes, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.1,
color="g")
axes.plot(train_sizes, train_scores_mean, 'o-', color="r",
label="Training score")
axes.plot(train_sizes, test_scores_mean, 'o-', color="g",
label="Cross-validation score")
axes.legend(loc="best")
return plt
def train_model(title, estimator):
cv = rmse_cv(estimator)
cv_error = cv.mean()
cv_std = cv.std()
# fit
estimator.fit(X_train, y_train)
# predict
y_train_pred = estimator.predict(X_train)
training_error = np.sqrt(mean_squared_error(y_train, y_train_pred))
train_r2 = r2_score(y_train, y_train_pred)
y_test_pred = estimator.predict(X_test)
test_error = np.sqrt(mean_squared_error(y_test, y_test_pred))
test_r2 = r2_score(y_test, y_test_pred)
# visualizing the result
df = pd.DataFrame({'Model':title, '(RMSE) CV Error': cv_error, 'CV Std': cv_std, '(RMSE) Training error':training_error, '(RMSE) Test Error':test_error, '(R2) Training Score':train_r2, '(R2) Test Score':test_r2}, index=[0])
return df
2.1 Linear Regression
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr_results = train_model('Linear Regression', lr)
lr_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | Linear Regression | 0.229457 | 0.016247 | 0.209983 | 0.217076 | 0.835861 | 0.817331 |
title = "Learning Curves"
estimator = lr
plot_learning_curve(estimator, title, X_var, y)
plt.show()

2.2 Random Forest Regressor
rf = RandomForestRegressor(max_depth = 6, random_state = 27)
rf_results = train_model('Random Forest Regressor [baseline]', rf)
rf_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | Random Forest Regressor [baseline] | 0.24443 | 0.015315 | 0.202519 | 0.234036 | 0.847323 | 0.787672 |
Hyperparameter Tuning
We will try to perfrom some hyperparameter tuning to see if we can improve our score on the test set
n_estimators = [int(x) for x in np.linspace(start = 100, stop=2000, num=20)]
max_features = [None, 'sqrt', 'log2']
max_depth = [np.arange(start = 3, stop = 16, step=1)]
max_depth.append(None)
min_samples_split = [2, 4, 5, 6, 7]
min_samples_leaf = [1, 2, 4]
bootstrap = [True, False]
random_grid ={'n_estimators': n_estimators,
'max_features': max_features,
'max_depth': max_depth,
'min_samples_split': min_samples_split,
'min_samples_leaf': min_samples_leaf,
'bootstrap': bootstrap}
rf_random = RandomizedSearchCV(estimator=rf, cv = 5, param_distributions = random_grid, random_state=27, n_jobs=-1, verbose=0)
rf_random.fit(X_train, y_train)
rf_random.best_params_
{'n_estimators': 1700,
'min_samples_split': 7,
'min_samples_leaf': 1,
'max_features': 'sqrt',
'max_depth': None,
'bootstrap': False}
best_random = rf_random.best_estimator_
rf_best_results = train_model('Random Forest [optimized]', best_random)
rf_best_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | Random Forest [optimized] | 0.217775 | 0.009606 | 0.103904 | 0.203305 | 0.959811 | 0.839772 |
title = "Learning Curves"
estimator = best_random
plot_learning_curve(estimator, title, X_var, y)
plt.show()

If we take a look at the curve we can see that the trend shows an increasing validation score, if we have more data we might be able to get it close to the training score. This is one of the downside of working with limited data on ensemble model.
2.3 XGBoost Regressor
xgb = XGBRegressor(verbosity = 0, random_state = 27)
xgb_results = train_model('XGBRegressor [baseline]', xgb)
xgb_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | XGBRegressor [baseline] | 0.224384 | 0.009016 | 0.071544 | 0.228214 | 0.980946 | 0.798104 |
d = {'Learning Rate':[],
'Mean CV Error': [],
'CV Error Std': [],
'Training Error': []}
for lr in np.arange(start= 0.05, stop=0.35, step=0.05):
xgb = XGBRegressor(n_estimators=500, learning_rate=lr, early_stopping=5)
cv_res = rmse_cv(xgb)
xgb.fit(X_train, y_train)
y_train_pred = xgb.predict(X_train)
d['Learning Rate'].append(lr)
d['Mean CV Error'].append(cv_res.mean())
d['CV Error Std'].append(cv_res.std())
d['Training Error'].append(np.sqrt(mean_squared_error(y_train, y_train_pred)))
xgb_tuning_1 = pd.DataFrame(d)
xgb_tuning_1
| Learning Rate | Mean CV Error | CV Error Std | Training Error | |
|---|---|---|---|---|
| 0 | 0.05 | 0.216038 | 0.011774 | 0.092352 |
| 1 | 0.10 | 0.225486 | 0.007819 | 0.058462 |
| 2 | 0.15 | 0.228645 | 0.005337 | 0.052823 |
| 3 | 0.20 | 0.227991 | 0.003771 | 0.051646 |
| 4 | 0.25 | 0.228967 | 0.006944 | 0.051581 |
| 5 | 0.30 | 0.231707 | 0.005858 | 0.051467 |
xgb_tuning_2 = pd.DataFrame(d)
xgb_tuning_2
print('Optimal parameter values are: ')
best = xgb_tuning_2.iloc[xgb_tuning_2.idxmin()['Mean CV Error']]
print('max_depth: {}'.format(int(best['max_depth'])))
print('min_child_weight: {}'.format(int(best['min_child_weight'])))
Optimal parameter values are:
max_depth: 3
min_child_weight: 1
d = {'max_depth':[],
'min_child_weight': [],
'Mean CV Error': [],
'CV Error Std': [],
'Training Error': []}
params2 = {'max_depth': list(range(3,10,2)), 'min_child_weight': list(range(1,6,2))}
for md in params2['max_depth']:
for mcw in params2['min_child_weight']:
xgb_model = XGBRegressor(n_estimators=1000, learning_rate=0.05, early_stopping=5, max_depth=md, min_child_weight=mcw )
cv_res = rmse_cv(xgb_model)
xgb_model.fit(X_train, y_train)
y_train_xgb = xgb_model.predict(X_train)
d['max_depth'].append(md)
d['min_child_weight'].append(mcw)
d['Mean CV Error'].append(cv_res.mean())
d['CV Error Std'].append(cv_res.std())
d['Training Error'].append(np.sqrt(mean_squared_error(y_train_xgb, y_train)))
n_estimators = np.arange(500, 1100, 100)
learning_rate = np.arange(0.05, 0.35, 0.05)
max_depth = np.arange(3,10,2)
min_child_weight = np.arange(1,6,2)
param_grid = dict(n_estimators=n_estimators, learning_rate=learning_rate, max_depth = max_depth, min_child_weight = min_child_weight)
xgb_random = RandomizedSearchCV(estimator=xgb, cv = kf, param_distributions = param_grid, scoring = 'neg_root_mean_squared_error', random_state=27, n_jobs=-1)
xgb_random.fit(X_train, y_train)
xgb_random.best_params_
{'n_estimators': 800,
'min_child_weight': 1,
'max_depth': 3,
'learning_rate': 0.05}
xgb_best = xgb_random.best_estimator_
xgb_best_results = train_model('XGBRegressor [optimized]', xgb_best)
xgb_best_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | XGBRegressor [optimized] | 0.208752 | 0.017149 | 0.133361 | 0.205716 | 0.933794 | 0.835949 |
title = "Learning Curves"
estimator = xgb_best
plot_learning_curve(estimator, title, X_var, y)
plt.show()

Based on the curve trend we can see that there is still potential for the training and validation/test score to improve given more data just like our Random Forest.
2.4 Gradient Boosting
gb = GradientBoostingRegressor(random_state = 27)
gbr_results = train_model('Gradient Boosting', gb)
gbr_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | Gradient Boosting | 0.220045 | 0.013427 | 0.172792 | 0.21274 | 0.888855 | 0.824556 |
Hyperparameter Tuning
learning_rate = np.arange(start= 0.05, stop=0.2, step=0.01)
n_estimators = np.arange(start = 100, stop = 2050, step=50)
max_depth = [x for x in np.linspace(1, 10, num=10)]
min_samples_split = [2, 4, 5, 6, 7]
min_samples_leaf = [1, 2, 4]
max_features = [None, 'sqrt', 'log2']
random_grid ={}
random_grid = {
'learning_rate':learning_rate,
'n_estimators': n_estimators,
'max_depth': max_depth,
'max_features': max_features,
'min_samples_split':min_samples_split,
'min_samples_leaf':min_samples_leaf
}
gb_random = RandomizedSearchCV(estimator=gb, cv = kf, param_distributions = random_grid, scoring = 'neg_root_mean_squared_error', random_state=27, n_jobs=-1)
gb_random.fit(X_train, y_train)
gb_random.best_params_
{'n_estimators': 650,
'min_samples_split': 6,
'min_samples_leaf': 1,
'max_features': 'log2',
'max_depth': 4.0,
'learning_rate': 0.07}
gb_best = gb_random.best_estimator_
gb_best_results = train_model('Gradient Boosting [optimized]', gb_best)
gb_best_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | Gradient Boosting [optimized] | 0.215909 | 0.013687 | 0.104709 | 0.197649 | 0.959186 | 0.848563 |
title = "Learning Curves"
estimator = gb_best
plot_learning_curve(estimator, title, X_var, y)
plt.show()

Similar to other ensemble model learning curve we can see an increasing trend in our validation score, given more data the accuracy might improve even more. But. even so this model yielded the highest accuracy so far.
2.5 Ridge Regression
ridge_alphas = [1e-15, 1e-10, 1e-8, 9e-4, 7e-4, 5e-4, 3e-4, 1e-4, 1e-3, 5e-2, 1e-2, 0.1, 0.3, 1, 3, 5, 10, 15, 18, 20, 30, 50, 75, 100]
ridge = Ridge(random_state = 27)
ridge_rob = make_pipeline(RobustScaler(), RidgeCV(alphas=ridge_alphas, cv=kf))
ridge_rob_results = train_model('Ridge Regression (rob)', ridge_rob)
ridge_rob_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | Ridge Regression (rob) | 0.230955 | 0.016572 | 0.211195 | 0.216493 | 0.833961 | 0.81831 |
ridge_norm = make_pipeline(MinMaxScaler(), RidgeCV(alphas=ridge_alphas, cv=kf))
ridge_norm_results = train_model('Ridge Regression (norm)', ridge_norm)
ridge_norm_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | Ridge Regression (norm) | 0.229402 | 0.015968 | 0.210444 | 0.216502 | 0.835139 | 0.818296 |
ridge_std = make_pipeline(StandardScaler(), RidgeCV(alphas=ridge_alphas, cv=kf))
ridge_std_results = train_model('Ridge Regression (std)', ridge_std)
ridge_std_results
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | Ridge Regression (std) | 0.229708 | 0.016488 | 0.21007 | 0.217024 | 0.835726 | 0.817418 |
title = "Learning Curves"
estimator = ridge_rob
plot_learning_curve(estimator, title, X_var, y)
plt.show()

pd.concat([lr_results, rf_best_results, xgb_best_results, gb_best_results, ridge_rob_results], axis=0, ignore_index=True)
| Model | (RMSE) CV Error | CV Std | (RMSE) Training error | (RMSE) Test Error | (R2) Training Score | (R2) Test Score | |
|---|---|---|---|---|---|---|---|
| 0 | Linear Regression | 0.229457 | 0.016247 | 0.209983 | 0.217076 | 0.835861 | 0.817331 |
| 1 | Random Forest [optimized] | 0.217775 | 0.009606 | 0.103904 | 0.203305 | 0.959811 | 0.839772 |
| 2 | XGBRegressor [optimized] | 0.208752 | 0.017149 | 0.133361 | 0.205716 | 0.933794 | 0.835949 |
| 3 | Gradient Boosting [optimized] | 0.215909 | 0.013687 | 0.104709 | 0.197649 | 0.959186 | 0.848563 |
| 4 | Ridge Regression (rob) | 0.230955 | 0.016572 | 0.211195 | 0.216493 | 0.833961 | 0.818310 |
Comparing all these different models we can see that XGBRegressor showed the best result with a CV error of 0.208752, the reason I picked this model even though Random Forest and Gradient Boosting showed a better test score and error is because we don’t want to select model based on our test performance, if we did that it would mean that our model overfit the test data since it may have a better performance due to chance alone. It’s best practice to perform any sort of model selection based on our CV error since it is more representative of our model generalization capability.
The performance on the test set should only then be use as a true out of sample performance metric once we’ve selected our model. Based on the difference in training and test performance we can also see that there is a high variance in our model due to the lack of data, this is further supported by the trend on the learning curve where the test score are still increasing alongside with the number of training data.
Exporting Model
import pickle
with open('travelio_apart.pickle','wb') as f:
pickle.dump(xgb_best,f)
import json
columns = {
'data_columns' : [col.lower() for col in X_var.columns]
}
with open("columns.json","w") as f:
f.write(json.dumps(columns))